Network biology approaches to interpret multi-omics data from gut organoids
Tamas Korcsmáros1,2
1 Earlham Institute, Norwich Research Park, Norwich, NR4 7UZ, UK
2 Quadram Institute, Norwich Research Park, Norwich, NR4 7UA, UK
Background
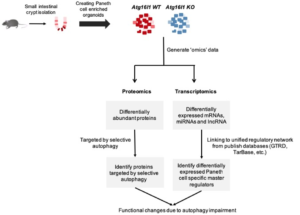
Autophagy maintains the intestinal homeostasis by allowing the epithelium to adapt to various stress situations, participating in antibacterial defense, controlling the microbiome composition and the immune response. Mutation in autophagy genes and deregulated autophagy are related to various human diseases including Crohn’s disease (CD) where autophagy impairment was shown to affect Paneth cells (PCs). Previously, we developed the Autophagy Regulatory Network resource (http://autophagyregulation.org) to better understand the mechanism and regulation of autophagy in disease pathomechanisms. To investigate autophagy-related processes and study potential master regulators in PCs, we combined ARN with multi-omics data from intestinal organoids. In particular, we investigated how damaged autophagy, often observed in CD, could affect the key cell functions of PCs.
Methods
We generated mouse intestinal organoids to compare the RNA expression and protein level of normal and autophagy impaired systems.
Results
We detected 284 proteins with altered protein levels by comparing the proteomic profiles of organoids derived from normal mice and mice with impaired autophagy. Our integrated analysis – combination of proteomics and network biology approaches – revealed autophagy-mediated mechanisms that degrade essential proteins belonging to key Paneth cell functions such as exocytosis, apoptosis and DNA damage repair. We performed validation experiments by generating full transcriptomics profiles of both organoid types. By mapping cell-type specific marker genes to the network derived from normal mice, we were able to identify regulators potentially contributing to Paneth cell specific functions. Among the seven putative master regulators, we identified four nuclear hormone receptors with links to inflammatory bowel disease, immunity and autophagy: Vdr, Rxra, Nr1d1 and Nr3c1. Subsequent analysis of the autophagy impaired mouse derived networks has enabled investigation of the effect of autophagy impairment on the regulatory landscape of Paneth cell.
Conclusion
We used both experimental and computational approaches to analyse and uncover the systems-level regulation of cellular processes dependent on autophagy in Paneth cells enriched organoids. Strikingly, the analysis revealed that when autophagy is impaired, nearly three hundred proteins display increased or decreased abundance, encompassing at least 18 functional processes. The established workflow will enable analysing human Paneth cells from clinical biopsies as well as use to investigate the regulatory effect of microbial challenges on Paneth cells in Crohn’s disease.