Course dates: November 4-8, 2019
Location: Gløshaugen campus, NTNU, Trondheim
Registration is closed
Recommended credits: 7,5 ECTS.
This course runs with a NTNU course code (BT8118). Note that in order to earn credits, you must be a student at NTNU, or apply for status as a visiting PhD student at NTNU by September 15. More information here: https://www.ntnu.edu/studies/researchcourses
Main organizer: Ines Heiland
Lecturers:
Ines Heiland (Department of Arctic and Marine Biology, UiT – The Arctic University of Norway)
Eivind Almaas (Department of Biotechnology and Food Science, NTNU)
Ronan Fleming (Luxembourg Center for Systems Biomedicine, University of Luxembourg)
Barbara Bakker (Faculty of Medical Sciences, University of Gronigen)
Schedule: tba
Preparations for the course:
- Read the following material: tba
- You should have Matlab installed on your laptop – version 2015a, 2015b, 2016a or 2016b, BUT NOT 2017a or 2017b.
- You must have Cobra toolbox installed on your laptop, please find the instructions here.
Description
This course will provide an introduction to the computational analysis and reconstruction of both small and genome-scale metabolic networks. The goal is to make students familiar with state-of-the-art computational tools and databases, as well as providing the students with hands-on experience. Furthermore, we will cover the mathematical basis of constraint-based analysis of genome-scale metabolic models and provide a foundation for stability and control analysis of dynamic models.
The first part of the course (2.5 days) will focus on the reconstruction and analysis of small scale, mechanistic models of metabolic pathways that allow the simulation of dynamic properties of the system. In this context, we will provide an introduction to stability and control analysis.
The second part of the course (2.5 days) will focus on the organization of large scale metabolic networks; their reconstruction and the mathematical basis of optimization approaches used in constraint-based modeling. We will use COBRApy, a Python based toolbox, for the analysis and manipulation of genome-scale metabolic models. Alternatively, the COBRA toolbox in MATLAB will be used. We will also discuss available methods for data integration.
During the course, a poster session will be organized. All students are expected to present their research projects during the poster session. This will provide an arena for the students to discuss potential application of modelling approaches to their own research with the lecturers of the course.
During the course, every student is expected to start a small, individual modelling project that will be the basis of the assessed project report. The aim is to apply the methods taught during the course to individual research projects ideally to be continued during their PhD.
Course program
The course will be given over one week (5 days, Monday through Friday), each day from 09-17 with a 1-hour lunch break. We will schedule lectures and practical exercises so that each will contribute about 50% to the intensive course week.
The students will receive a reading list before the course and will develop a course project during the course week. They are furthermore expected to bring a poster about their own research projects to be presented in a poster session during the course. The project report will require the students to apply methods taught during the course, and the students will have to prepare and return the report 4 weeks after the end of course teaching.
Prerequisites
Familiarity with Linux and a programming language (preferably Python) is expected. Basic knowledge in biochemistry, linear algebra and statistics are required.
Learning outcomes and competence
- Explain the organization and structure of metabolic models
- Knowledge about relevant databases
- Capable of reconstructing metabolic models
- Explain the underlying principles of constraint based modeling approaches
- Explain stability and control analysis
- Proficiency in use of COBRA toolbox for genome-scale metabolic modeling
- Explain limits of small and genome-scale metabolic analysis
- Knowledge about tools for data integration
Evaluation
Poster, Exercises and project report (10 pages) that will be evaluated. Grades: pass/fail
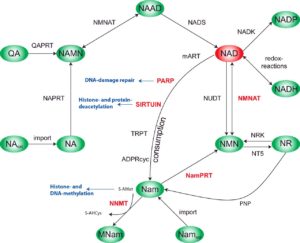
Illustration: Ines Heiland, DOI: 10.1042/BST20150133